The power of genetic medicine for rare immune diseases
 In the past few months, people around the world have adopted new habits to protect themselves from a novel virus. Yet for people with primary immunodeficiency, born with a compromised immune system, reducing the risk of infection is nothing new: they have been more vulnerable to viruses and bacteria all of their lives.
In the past few months, people around the world have adopted new habits to protect themselves from a novel virus. Yet for people with primary immunodeficiency, born with a compromised immune system, reducing the risk of infection is nothing new: they have been more vulnerable to viruses and bacteria all of their lives.
For World Primary Immunodeficiency Week, we have talked to one of St Vincent's Clinic Foundation's research grant recipients, A/Prof Tri Giang Phan, to learn how he aims to improve diagnosis and treatment of primary immunodeficiency diseases by combining the power of genomic medicine and data science.
In the medical community, an old saying goes « when you hear hoof beats, think horses, not zebras ». This means that when making a diagnosis, doctors should focus on the likeliest possibilities. Unfortunately, this does not apply to primary immunodeficiency disorders. Because they are so rare, these immune diseases are considered the ‘zebras’ of the medical world. As part of CIRCA (Clinical Immunogenomic Research Consortium Australia), A/Prof Tri Giang Phan is looking for the equivalent of ‘stripes’ - or rather, the genetic cause of those stripes.
Can you tell us why this research is so important?
First of all, many patients with primary immunodeficiency are extremely sick. Most of them are children, and some suffer from life-threatening infections that you and I would have no trouble combating. Unfortunately, some of them spend their lives in and out of intensive care. On top of that, it can take 5 to 10 years for these patients to be properly diagnosed. When we do find a diagnosis, we are able to start them on a specific treatment to correct their gene defect, and this can have a tremendous impact on their lifespan and their quality of life.
Secondly, this research has the potential to have impacts way beyond patients with rare immune diseases. As our patients have a dysfunctional immune system, it tends to overact, which causes autoimmune diseases and allergies. By studying these patients and understanding the genetic cause of their immune disorder, we expect to discover new ways to treat these more common diseases. A better understanding of the immune system will also help us drive the development of new immunotherapies to treat certain types of cancer.
How do you proceed to find the genetic causes of these immune diseases?
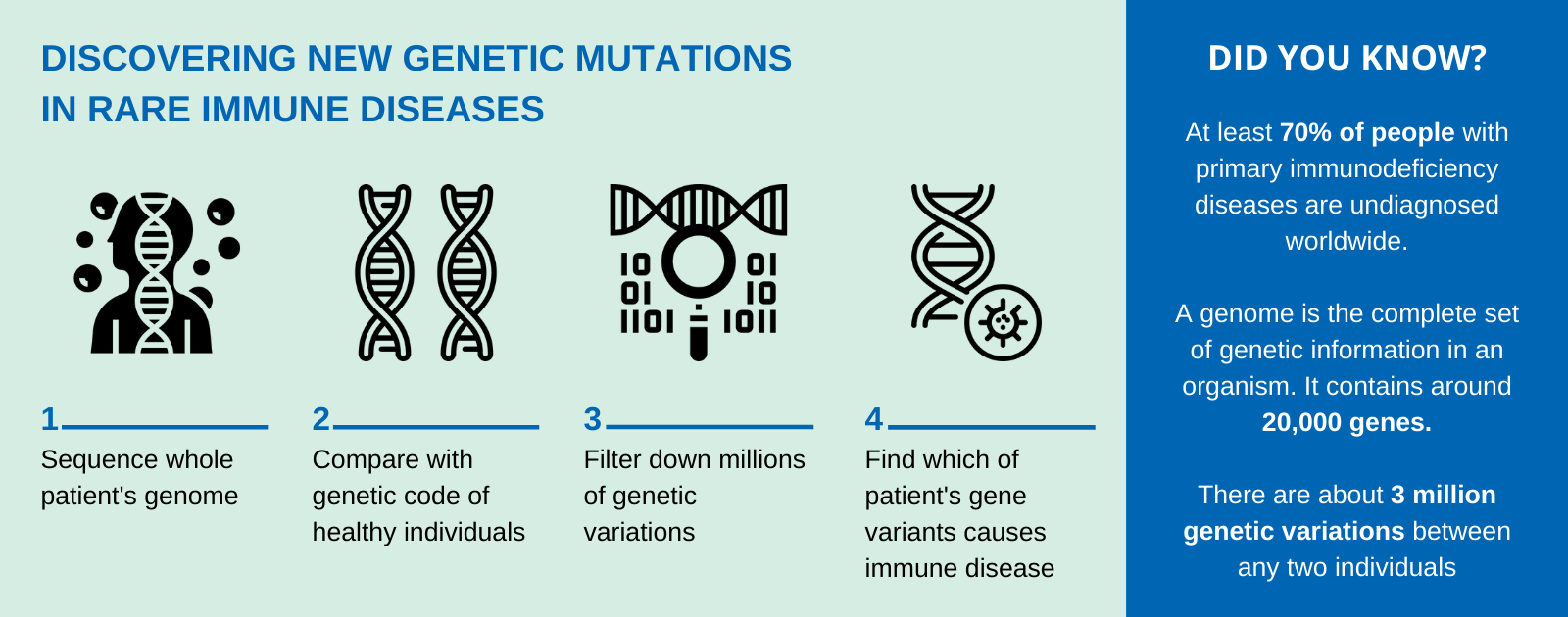
The diagnosis is no small task. It involves sequencing the whole genome of a patient, comparing it with the genetic code of healthy individuals to look for differences, and filtering these variants to find out which gene mutation is at the root-cause of the immune disease. There are about 3 million variants between any two individuals. Many of them do not have any consequences or are what make us unique (like eye or hair colour) and are not relevant for our investigation, so it is a bit like finding a needle in a haystack.
To cut through the amount of data, we started by analysing a portion of the genome only, focusing on 300 genes (out of 20,000). This “shortcut” enabled us to streamline our diagnostic process and find the genetic cause of disease for 40% of our patients with primary immunodeficiency.
Now our goal is to help the remaining 60% of our patients. Our take is that their immune defect is in a part of the genome we have not yet analysed. It means we need to dig deeper into the genetic code and re-analyse it in its entirety, with no shortcuts this time.
Is this where the St Vincent’s Clinic Foundation grant comes in?
Exactly. This re-analysis project entails multiple components, and the St Vincent’s Clinic Foundation grant is funding the very beginnings of our work.
Dr Leslie Burnett at the Kinghorn Centre for Clinical Genomics, who is co-investigator on the grant, has developed a Machine Learning tool to analyse a genome sequence, which can run the analysis within 2 to 3 weeks, as opposed to the 6 to 9 months with a trained bioinformatician and genetic pathologist. It has tremendously accelerated our throughput. It will allow us to scale our work and look beyond the 300 genes that we have analysed so far - and find the gene mutations in our remaining patients.
From there, what the grant is allowing us to do is to develop and run novel bioinformatic tools. These tools, which combine biology and computer science, are extremely valuable to analyse and interpret data sets as large and complex as genomic sequences. Complementing our Machine Learning system, these bioinformatic tools will enable us to efficiently mine the patients’ genomic data and help us work our way through the millions of genetic variants. We expect these additional tools will help us narrow down those variants to half a dozen, and thus accelerate our diagnosis process. A trained bio-mathematician will then look through the patient’s genes singled out by the tools, and see if one of them is ‘defective’ and causing the immune disease.
Once a disease-causing mutation is uncovered, the project will also involve additional experimentation to prove it is indeed the case. This research relies on funding from several sources, and the St Vincent’s Clinic Foundation grant is a very important part of that funding as it allows us to take this multi-pronged approach to solve this.
How has the COVID-19 crisis impacted your research project?
Luckily, the bulk of our research work is done on a computer, so we have been able to carry on. The team has been meeting regularly online, but I have to admit, this is not as efficient as being in the same room.